Langevin Sampling
Intro
Langevin sampling is another sampling technique that just requires that you can compute . Specifically, it constructs an Stochastic Differential Equation (SDE) such that the stationary distribution is . Lets look into the theory of how a random walk can sampling a distribution.
Itô Calculus and Fokker-Plank Equation
Let be Brownian motion. So , and . Consider . This isn’t the usual calculus definition of a change of a function, in fact it can be shown that doesn’t exist, and it makes sense intuitively, its a distribution, so it won’t converge to some exact value. But Itô made an interesting observation.
Where, are equidistant points from to What this tells us is that . This leads us to Itô’s lemme which states that for
Where follows a stochastic process, which is why doesn’t vanish. You can derive this by doing a Taylor expansion and then removing the terms that vanish. Lets specify the stochastic process for :
substituting into the expression for yields:
and after taking the expectation over which is based on the true distribution of at time , The terms with disappear and after doing some algebra we get the Fokker-Plank Equation. I like the proof in this article if you want to look at the details.
Fokker-Plank Equation
So ideally we want to converge to which is the original distribution we wanted to sample from. So we can agree that as time limits, then the partial derivative with respect to time should equal , which would imply that the distribution converges. It can be shown that the following stochastic process does have our desired stationary distribution.
I recommend this well written article to see why that is the case.
Implementation
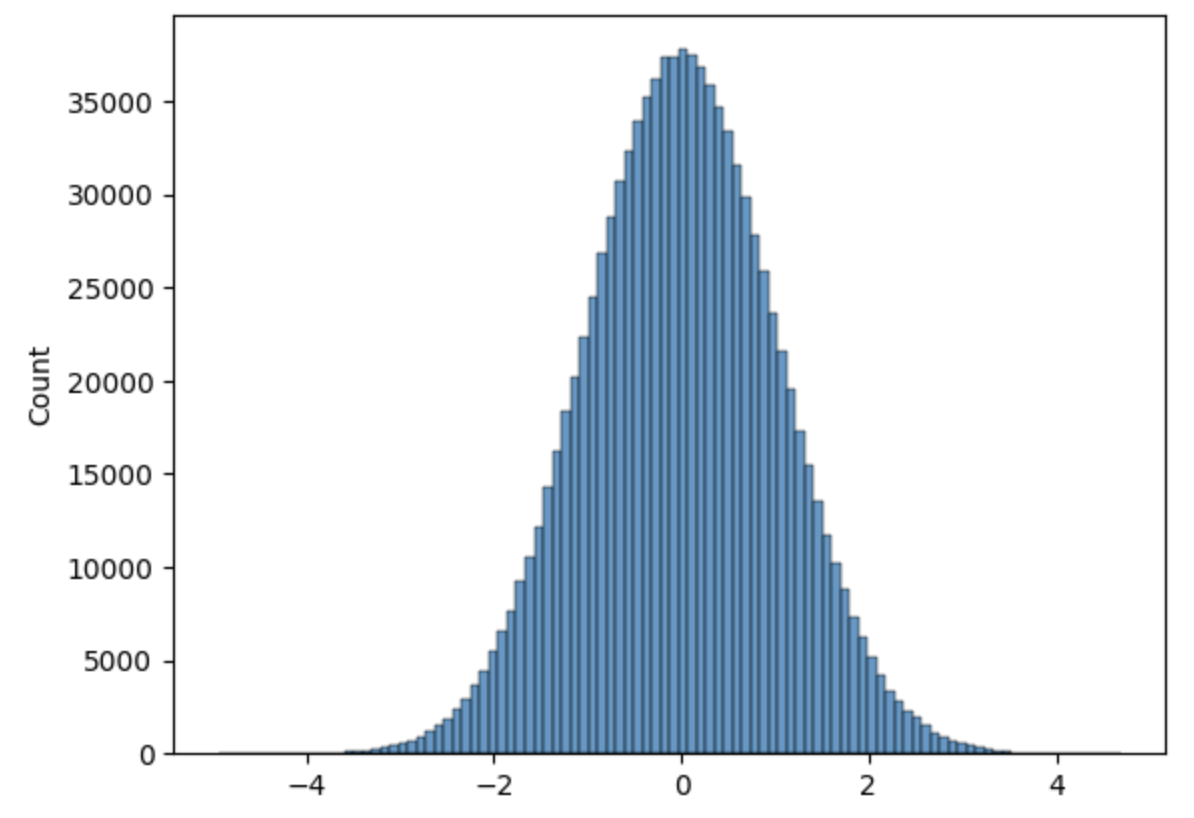
Here is a short code to sample a standard normal distribution.
import matplotlib.pyplot as plt
import numpy as np
import math
def pdf(x, mu, sigma):
# gaussian pdf
return (1/(sigma*math.sqrt(2*math.pi))) * math.exp((-1/2)*((x-mu)/sigma)**2)
def pdf_deriv(x, mu, sigma):
return (-1/((sigma**3)*math.sqrt(2*math.pi))) * math.exp((-1/2)*((x-mu)/sigma)**2) * (x-mu)
# take on step of the Monte Carlo Process
def step(x, a, mu, sigma):
# taking the gradient of ln(pdf) = 1/pdf * pdf_deriv
return x + a * (1/pdf(x, mu, sigma))*pdf_deriv(x, mu, sigma) + math.sqrt(2*a)*np.random.normal(0,1,1)[0]
x_vals = []
x = 1
alpha = 0.1
num_iter = 1000000
for i in range(num_iter):
x_vals.append(x)
x = step(x, alpha, 0, 1)
plt.hist(x_vals, bins=100)
plt.show()
References and Resources
https://www.thphys.uni-heidelberg.de/~wolschin/statsem21_6s.pdf
https://abdulfatir.com/blog/2020/Langevin-Monte-Carlo/
https://www.youtube.com/watch?v=Z5yRMMVUC5w&list=PLUl4u3cNGP63ctJIEC1UnZ0btsphnnoHR&index=17