Distances Between Subspaces
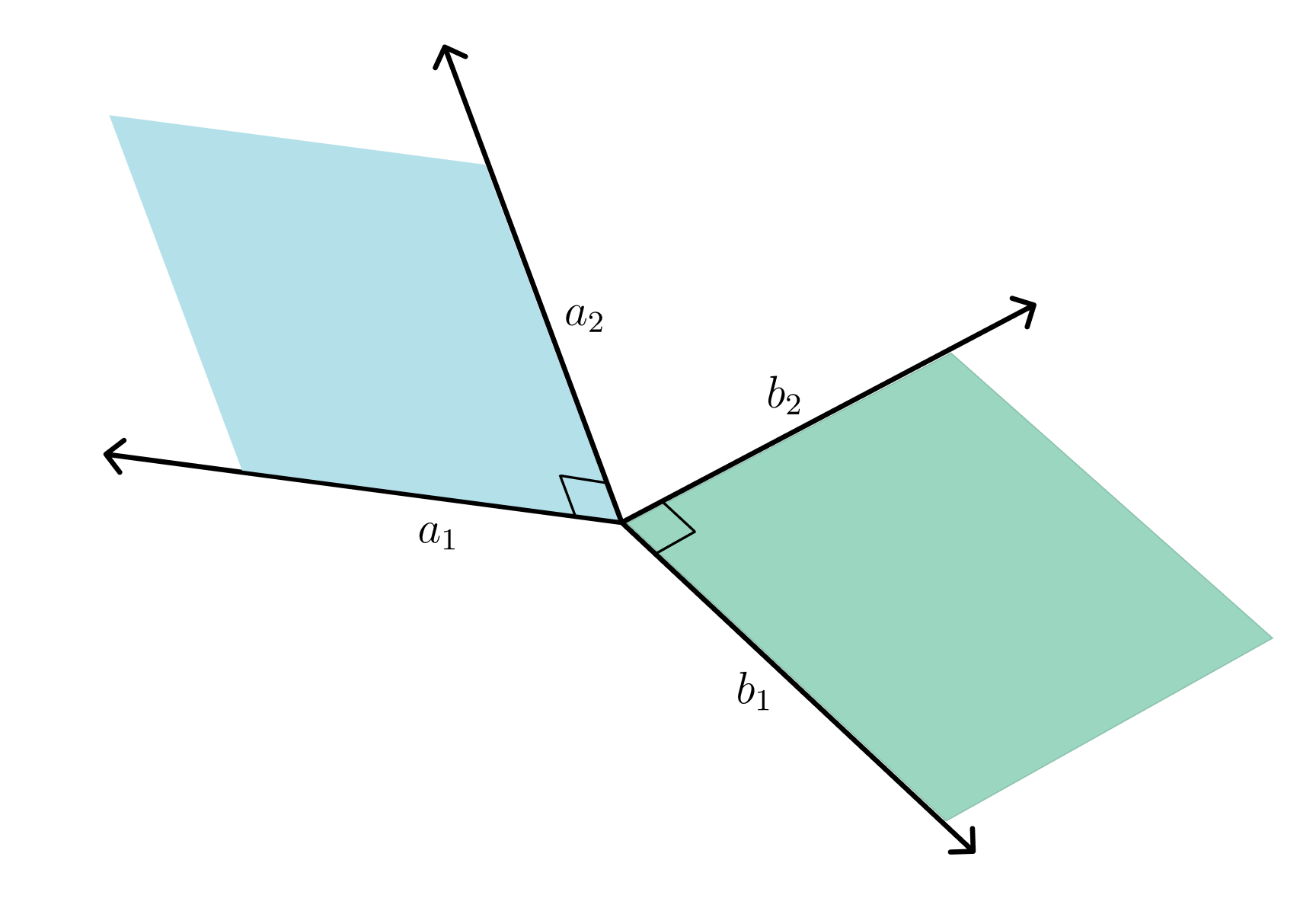
This question came up when trying to compare kernels as well as how low dimensional projections change over time. Intuitively it isn’t clear how to create a distance metric between subspaces. For example, lets say we are in , and we have to subspaces . To this end, I will try to explain the Grassman distance.
Principle Angles
Then the idea of principle angles is to iteratively find vectors in that have the closest angle. More formally, at iteration , we have the following optimization problem:
We keep repeating this procedure for following iterations with the additional constraint that if we are iteration , then and . Therefore, we are trying to progressively find the most similar orthonormal vectors in and .
Let be orthonormal basis for respectively. Then from our previous iterative algorithm, let , so we want to maximize . Let’s compute the SVD of . Substituting yields, . Therefore, if we align with the top singular vector in and with the top singular vector in , then the resulting value will be the largest singular value . In fact the singular values are the ordered solutions to the aforementioned iterative optimization procedure (think about why that is the case). The Grassman metric is now defined as:
I am not going to prove why the triangle inequality holds for this to be a metric, but you can look at some of the resources I linked below. I might revisit it and add it to the blog later.
Code
import torch
import math
def grassman(A, B):
# get orthogonal basis
Q_A, R = torch.linalg.qr(A)
Q_B, R = torch.linalg.qr(B)
# get singular values
_, S, _ = torch.svd(Q_A.T @ Q_B)
# cosine inverse
thetas = torch.arccos(S)
# return norm
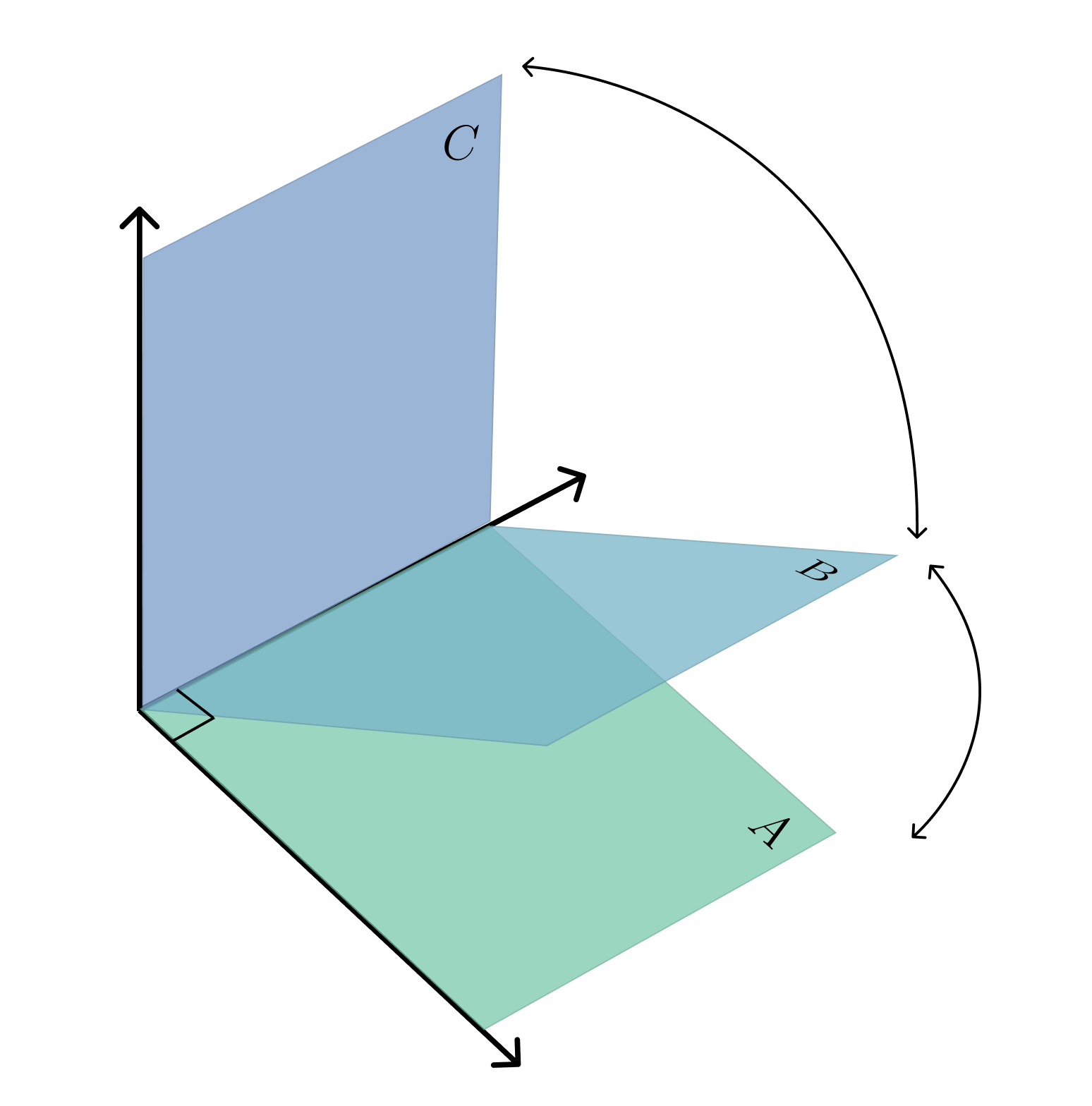
return torch.norm(thetas, p=2)A = torch.tensor([
[1, 0],
[0, 1],
[0, 0]
]).float()
B = torch.tensor([
[math.sqrt(0.5), 0],
[0, 1],
[math.sqrt(0.5), 0]
]).float()
C = torch.tensor([
[0, 0],
[1, 0],
[0, 1]
]).float()
>>> grassman(A,B)
tensor(0.7854)
>>> grassman(A,C)
tensor(1.5708)
>>> grassman(C,B)
tensor(0.7854)
References
https://en.wikipedia.org/wiki/Grassmannian
https://kristianeschenburg.netlify.app/post/comparing-subspaces/
https://web.ma.utexas.edu/users/vandyke/notes/deep_learning_presentation/presentation.pdf
https://galton.uchicago.edu/~lekheng/work/schubert.pdf